Pooling in a predictive model of V1 explains functional and structural diversity across species
Abstract
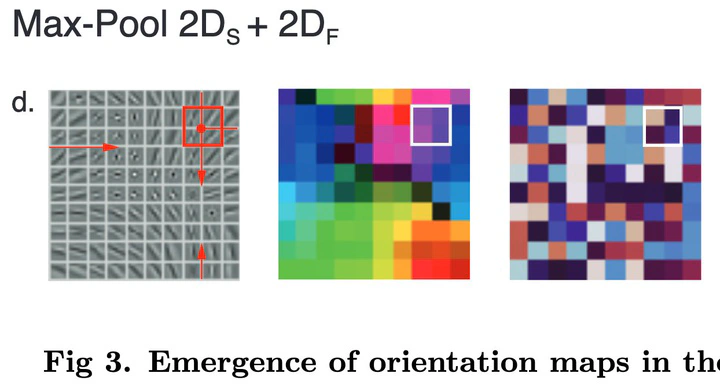
Neurons in the primary visual cortex are selective to orientation with various degrees of selectivity to the spatial phase, from high selectivity in simple cells to low selectivity in complex cells. Various computational models have suggested a possible link between the presence of phase invariant cells and the existence of cortical orientation maps in higher mammals’ V1. These models, however, do not explain the emergence of complex cells in animals that do not show orientation maps. In this study, we build a model of V1 based on a convolutional network called Sparse Deep Predictive Coding (SDPC) and show that a single computational mechanism, pooling, allows the SDPC model to account for the emergence of complex cells as well as cortical orientation maps in V1, as observed in distinct species of mammals. By using different pooling functions, our model developed complex cells in networks that exhibit orientation maps (e.g., like in carnivores and primates) or not (e.g., rodents and lagomorphs). The SDPC can therefore be viewed as a unifying framework that explains the diversity of structural and functional phenomena observed in V1. In particular, we show that orientation maps emerge naturally as the most cost-efficient structure to generate complex cells under the predictive coding principle.

this paper follows this COSYNE presentation :
(2020). Modelling Complex-cells and topological structure in the visual cortex of mammals using Sparse Predictive Coding. Computational and Systems Neuroscience (Cosyne) 2020.
see a related work describing SDPC in:
- more about the role of top-down connections: